LDPAC
LDPAC is a method for detecting spurious associations using p-values.
The strength of the method is its simplicity; it requires no other information
but p-values.
We provide both the Web server
where user can upload p-values,
and the Software binary.
Our method uses internally stored HapMap Phase III data to retrieve LD information.
USAGES
- Quick check: just drop 100 p-values around a suspicious association into our web server. You can instantly check if the association is spurious.
- Genome-wide check: since LDPAC is so fast, you can put whole genome p-values into one file and check spurious associations in hours of time.
- Resque: our method can detect possibly true associations from discarded associations by QC, if QC was too harsh.
HOW IT WORKS
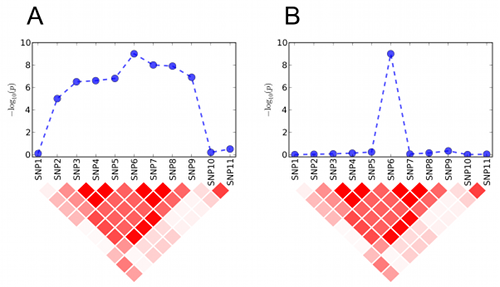
The intuition is that if the association is caused
by a true genetic effect, neighboring markers in LD should show
comparably significant p-values (Figure A). If not, it may be evidence of
spurious association (Figure B). Previous studies have applied the same
idea but only manually without a formal statistical framework.
Our method LDPAC provides a systematic framework
to quantitatively measure the evidence of spurious associations.
EXAMPLE
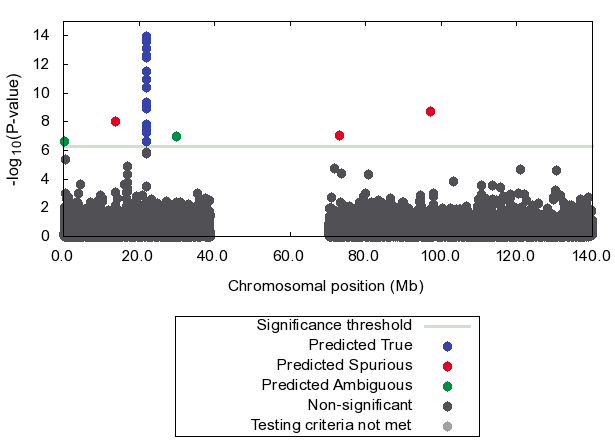
We take WTCCC CAD chr 9 data
where a confirmed association exists.
We put 5 spurious associations back into the data
which were originally excluded by QC in WTCCC study.
Figure shows that our method effectively detects 3
out of 5 spurious associations (red dots)
using only p-values.
The true association is correctly predicted as true (blue dots).
FUNDING INFORMATION
B.H. and E.E. are supported by National Science Foundation grants 0513612, 0731455, 0729049 and 0916676, and NIH grants K25-HL080079 and U01-DA024417. B.H. is supported by the Samsung Scholar- ship. This research was supported in part by the University of California, Los Angeles subcontract of contract N01-ES-45530 from the National Toxicology Program and National Institute of Environmental Health Sciences to Perlegen Sciences.

